Stat 414 – Review 1 Problem Solutions
1) Data was collected on corn grown on the island of Antigua (corn.txt). The response variable is the harvest weight (harvwt) per plot (unknown units). There are eight sites with eight separate plots within each site where the corn is grown under the same treatment condition. One question of interest is whether site has an effect on harvest weight.
(a) Identify the response variable and the Level 1 units and Level 2 units.
Response variable = harvest weight
Level 1 units = plots
Level 2 units = sites
(b) Produce a graph of the harvest weights for the eight sites. Does there appear to be substantial site to site variation?
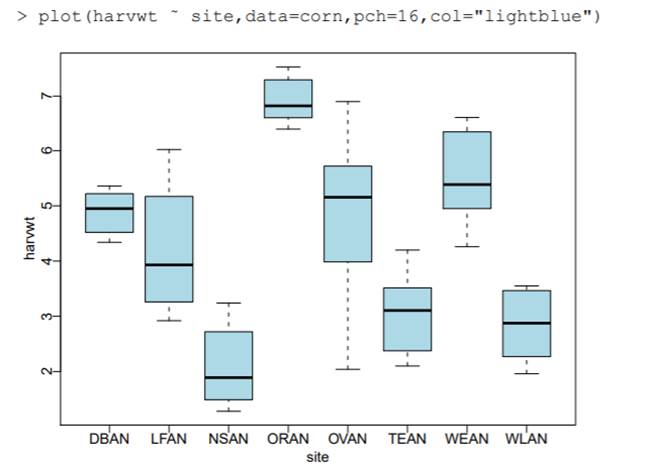
Yes, we do see
substantial site to site variation, for example the ORAN site and NSAN site
indicate very different havest weight values.
(c) Explain how we could use a traditional one-way ANOVA to model these data: identify the parameters, identify the error. Write out the statistical model (with proper indices, assumptions of error distribution, parameters).
Yij = harvest weight of ith plot in
site j
Yij = ![]() +
+ ![]() j + eij j = 1, …, 8 sites, eij ~ N(0,
j + eij j = 1, …, 8 sites, eij ~ N(0, ![]() 2)
2)
where ![]() is the overall mean
harvest weight in the population,
is the overall mean
harvest weight in the population, ![]() j is the fixed effect of the jth site, and
j is the fixed effect of the jth site, and ![]() 2 is
the within site variation (or the plot to plot variation, which we are assuming
is the same for each site). For one-way ANOVA the
2 is
the within site variation (or the plot to plot variation, which we are assuming
is the same for each site). For one-way ANOVA the ![]() j are considered fixed effects; so these are the
“effects” (not the group means, think effect coding).
j are considered fixed effects; so these are the
“effects” (not the group means, think effect coding).
(d) State null and alternative hypothesis and report a test statistic and p-value for assessing the site-to-site variation. Also produce a table of the estimated coefficients for the sites.
H0: ![]() 1 = …. =
1 = …. = ![]() 7 = 0
7 = 0
Ha: at least one ![]() j ≠ 0
j ≠ 0
(So we only need 7 of them and we
compare the set to zero, not just each other)
F = 17.4, df = 7 and 24, p-value < .0001. We have strong evidence that the population
mean weights are these sites are not equal.
|
> summary(lm(harvwt~site, contrasts=list(site="contr.treatment"))) This gives indicator
parameterization Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.8850
0.3800 12.856 2.96e-12 *** siteLFAN -0.6775
0.5374 -1.261 0.219513 siteNSAN -2.7950 0.5374
-5.201 2.50e-05 *** siteORAN 2.0300 0.5374
3.778 0.000922 *** siteOVAN -0.0525 0.5374
-0.098 0.922984 siteTEAN -1.8487 0.5374
-3.440 0.002135 ** siteWEAN 0.6412 0.5374
1.193 0.244412 siteWLAN -2.0438 0.5374
-3.803 0.000865 *** |
> summary(lm(harvwt~site, contrasts=list(site="contr.sum"))) This gives effect
parameterization Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 4.29172
0.13434 31.946 < 2e-16 *** site1 0.59328 0.35544
1.669 0.10808 site2 -0.08422 0.35544
-0.237 0.81471 site3 -2.20172 0.35544
-6.194 2.12e-06 *** site4 2.62328 0.35544
7.380 1.28e-07 *** site5 0.54078 0.35544
1.521 0.14121 site6 -1.25547 0.35544
-3.532 0.00170 ** site7 1.23453 0.35544
3.473 0.00197 ** |
|
By the way, there is a
nice “display()” function in the arm package. But
these are more about effects or group differences rather than estimates for
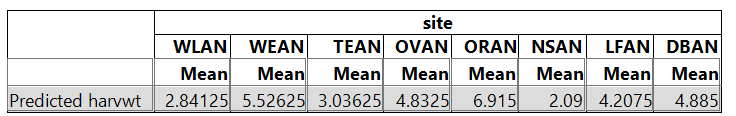
the sites. > sapply(split(fitted(lm(harvwt~site)), site),mean) DBAN
LFAN NSAN ORAN
OVAN TEAN WEAN
WLAN 4.88500 4.20750 2.09000
6.91500 4.83250 3.03625 5.52625 2.84125 |
|
|
So
these do match the group means. |
|
|
|
|
|
|
|
Now suppose we want to use a multilevel model. The main advantages are the ability to have plot level covariates, as well as site level covariates and an estimate of the error associated with the site level.
(e) Write out a statistical model for such a multilevel model, defining the parameters, with proper indices, assumptions of error distributions.
Yij = harvest weight of ith
plot in site j
Yij = ![]() +
+ ![]() j + eij j = 1, …, 8 sites, eij ~ N(0,
j + eij j = 1, …, 8 sites, eij ~ N(0, ![]() 2) and
2) and ![]() j ~ N(0,
j ~ N(0, ![]() 2)
2)
where ![]() is the overall mean harvest
weight in the population,
is the overall mean harvest
weight in the population, ![]() is the site
to site variability, and
is the site
to site variability, and ![]() 2 is
the within site variation (or the plot to plot variation, which we are assuming
is the same for each site).
2 is
the within site variation (or the plot to plot variation, which we are assuming
is the same for each site).
(f) State null and alternative hypothesis and report a test statistic and p-value for assessing the site-to-site variation.
H0: ![]() = 0
= 0
Ha: ![]() > 0
> 0
|
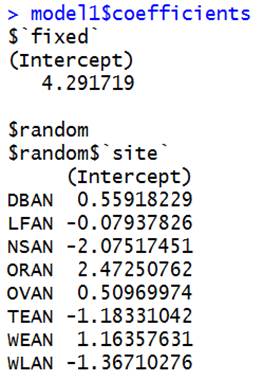
> model1=lme(fixed=harvwt~ 1, random =
~1 | site) > model2=gls(harvwt~1) > anova(model1,
model2) Model df AIC
BIC logLik Test
L.Ratio p-value model1 1
3 100.4163 104.7183 -47.20816 model2 2
2 126.4154 129.2834 -61.20772 1 vs 2 27.99912 <.0001 |
(g) Also produce a table of the estimated coefficients for the sites. How does these compare to the estimates from (d)? Why?
|
DBAN is first site, 4.291719 + .55918 = 4.851 |
> model1$fitted fixed site 1 4.291719 4.850901 2 4.291719 4.212340 3 4.291719 2.216544 4 4.291719 6.764226 5 4.291719 4.801418 6 4.291719 3.108408 7 4.291719 5.455295 8 4.291719 2.924616 9 4.291719 4.850901 10 4.291719 4.212340 11 4.291719 2.216544 12 4.291719 6.764226 13 4.291719 4.801418 14 4.291719 3.108408 15 4.291719 5.455295 16 4.291719 2.924616 17 4.291719 4.850901 18 4.291719 4.212340 19 4.291719 2.216544 20 4.291719 6.764226 21 4.291719 4.801418 22 4.291719 3.108408 23 4.291719 5.455295 24 4.291719 2.924616 25 4.291719 4.850901 26 4.291719 4.212340 27 4.291719 2.216544 28 4.291719 6.764226 29 4.291719 4.801418 30 4.291719 3.108408 31 4.291719 5.455295 32 4.291719 2.924616 |
> sapply(split(fitted(model1),
site),mean)
DBAN
LFAN NSAN ORAN
OVAN TEAN WEAN
WLAN
4.850901 4.212340 2.216544
6.764226 4.801418 3.108408 5.455295 2.924616
DBAN decreased
from 0.593 to 0.559. All of the
estimated effects got closer to zero because of the “shrinkage” that occurs
when we treat them as random effects. (So the estimated group means shrink
towards 4.2819 – for example DBAN decreases from 4.885 to 4.851, WLAN increased
from 2.841 to 2.925.
(h) Estimate and interpret the intraclass correlation coefficient. (How does this compare to R2 from the one-way ANOVA?)
|
|
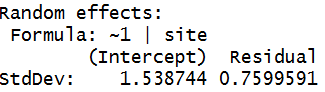
ICC = 2.3677/2.94527 = .8039, about
80% of the variation in response is at the site level. It’s
a little lower because we are now assuming more variation in the data to be
explained (MSTotal for the fixed model is 2.176).
This is in the ball park of R2
(i) Examine the residuals vs. fits graphs for this model and summarize what you learn.
|
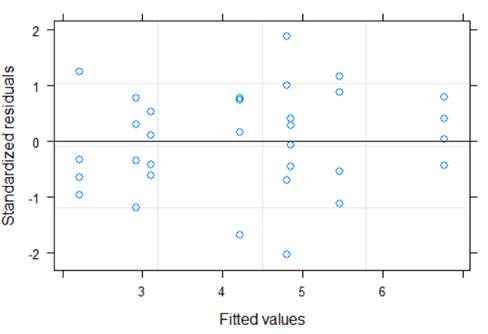
plot(model1)
|
There does not appear to be huge violations in terms on linearity (though not really a condition in this one-way anova type model but don’t see any concerning patterns) or equal variance.
Below are the standard deviations of the residuals per level. We do have some that are more than twice the size of others (e.g., OVAN and DBAN).

2) A researcher is interested in seeing how different light conditions (Long = 16 hr days, Short = 8 hr days) affects the concentration of an enzyme in golden hamsters in heart tissue. Ten hamsters are randomly placed into each of the two light treatment groups, for a total of twenty hamsters. After a treatment period of several weeks, the hamsters are sacrificed and enzyme concentration (mg/ml) is measured in four subsamples of heart tissue for each hamster. The data is in the file hamster.txt. The primary research question is to compare the effects of light on the enzyme concentration in heart tissue.
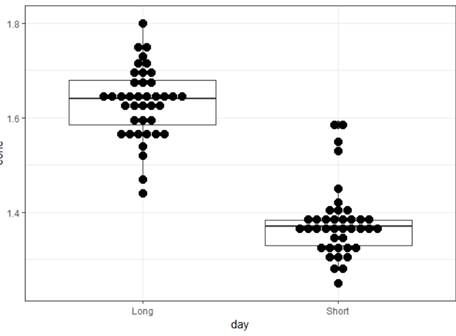
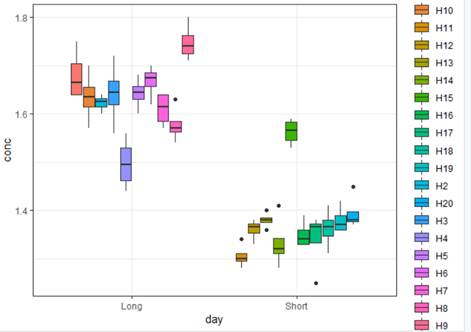
(a) Display a graph that highlights the primary research question.
Clearly the long day is associated with higher concentrations of the heart enzyme. There is variation in the measurements within each hamster, but that is smaller than the treatment to treatment variation. The within group variation in the hamsters looks pretty similar.
ggplot(hamsters, aes(y=conc, x = day, fill = id)) +
geom_boxplot()+
# geom_dotplot(binaxis='y',
stackdir='center', dotsize=1)
+
theme_bw()
(b) Fit and interpret a model that addresses the primary research question. Summarize your results.
Fitting a linear
model between concentration and day with hamsters as a random effect (following
a normal distribution). (Note: You could consider using a no intercept model
here as there should not be any concentration on day 0.)
|
> model1= lmer(conc ~ day + (1 | id)) > summary(model1) Linear mixed model fit by
REML ['lmerMod'] Formula: conc
~ day + (1 | id) REML criterion at
convergence: -227.7 Scaled residuals: Min
1Q Median 3Q
Max -2.30009 -0.54445
-0.03795 0.54010 1.93376 Random effects: Groups
Name Variance Std.Dev. id
(Intercept) 0.004216 0.06493 Residual 0.001644 0.04055 Number of obs:
80, groups: id, 20 Fixed effects: Estimate Std. Error t value (Intercept) 1.63225
0.02151 75.884 dayShort -0.25550 0.03042
-8.399 |
The t-statistic for dayShort is -0.255 /
0.0304 = -.12775/.01521 = -8.4, which is
large (I consider anything larger than 2 in absolute value to be large), giving
us convincing evidence that, after adjusting for the hamster to hamster
variation, the mean concentration increases with time.
With indicator parameterization, the intercept predicts the mean
concentration for the Long day treatment to be 1.63225 (with standard error
0.02151) or 1.37675 for the Short day treatment for the average hamster.
The dayShort slope estimates that the average
concentration with the Short day treatment will be 0.2555 lower than average Long
day treatment concentration for the average hamster.
An approximate 95% confidence interval for the difference (long
minus short) is -.255 + 2(.030) or (0.195, 0.315).
This was a randomized experiment, so we can conclude that the Long
treatment causes an increase in the concentration of the heart enzyme.
The hamsters explain .004216/(.004216 + .001644) or 71.9% of the random
variation in the concentrations, after adjusting for the treatments.
3)
(a) A has the largest ICC because the
variation between the schools is the largest and B has the smallest ICC.
(b) The estimated effect would increase because there would be
less shrinkage to the overall mean due to the larger sample size.
(c) The width of the confidence interval would decrease because it
would be a more precise estimate.
4)
(a)
90.86
(b) 90.86 – sqrt(10.66)
(c) 90.86 + 1.4145 + 2 × sqrt(10.66)
(d) 90.86 + 3.113